Efficient polymer-mediated delivery of gene-editing RNP payloads
Professor Theresa Reineke’s research on genome-editing payloads continues. An article on the latest break-through research, Efficient Polymer-Mediated Delivery of Gene-Editing Ribonucleoprotein Payloads through Combinatorial Design, Parallelized Experimentation, and Machine Learning, has been published in ACS Nano.
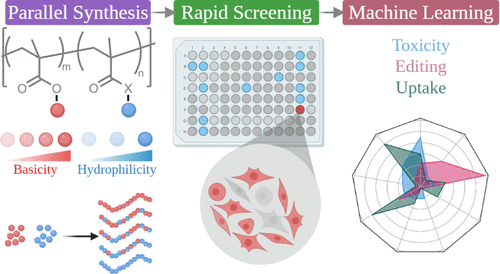
According to the abstract, "Chemically defined vectors such as cationic polymers are versatile alternatives to engineered viruses for the delivery of genome-editing payloads. However, their clinical translation hinges on rapidly exploring vast chemical design spaces and deriving structure–function relationships governing delivery performance. Here, we discovered a polymer for efficient intracellular ribonucleoprotein (RNP) delivery through combinatorial polymer design and parallelized experimental workflows. A chemically diverse library of 43 statistical copolymers was synthesized via combinatorial RAFT polymerization, realizing systematic variations in physicochemical properties. We selected cationic monomers that varied in their pKa values (8.1–9.2), steric bulk, and lipophilicity of their alkyl substituents. Co-monomers of varying hydrophilicity were also incorporated, enabling elucidation of the roles of protonation equilibria and hydrophobic–hydrophilic balance in vehicular properties and performance. We screened our multiparametric vector library through image cytometry and rapidly uncovered a hit polymer (P38), which outperforms state-of-the-art commercial transfection reagents, achieving nearly 60% editing efficiency via nonhomologous end-joining. Structure–function correlations underlying editing efficiency, cellular toxicity, and RNP uptake were probed through machine learning approaches to uncover the physicochemical basis of P38’s performance. Although cellular toxicity and RNP uptake were solely determined by polyplex size distribution and protonation degree, respectively, these two polyplex design parameters were found to be inconsequential for enhancing editing efficiency. Instead, polymer hydrophobicity and the Hill coefficient, a parameter describing cooperativity-enhanced polymer deprotonation, were identified as the critical determinants of editing efficiency. Combinatorial synthesis and high-throughput characterization methodologies coupled with data science approaches enabled the rapid discovery of a polymeric vehicle that would have otherwise remained inaccessible to chemical intuition. The statistically derived design rules elucidated herein will guide the synthesis and optimization of future polymer libraries tailored for therapeutic applications of RNP-based genome editing.”
In addition to Reineke, authors were Ramya Kumar, Ngoc Le, Zhe Tan, Mary Brown, and Shan Jiang.