IT researchers use computer models to unravel the inner workings of the cell
New findings by University of Minnesota biomedical engineering researchers are now helping to better understand how cells sense their mechanical environment, and how cells divide. The research could provide clues to controlling stem cell development and fighting cancer.
Controlling stem cell fate has been accomplished largely by biochemical means. However, recent studies indicate that stem cells can also be controlled by changing the mechanical stiffness of their environment, with neurons resulting on soft surfaces, muscle cells resulting on semi-stiff surfaces and bone cells resulting on stiff "glass-like" surfaces.
David Odde, professor of biomedical engineering in the university's Institute of Technology, and his team of biomedical engineering Ph.D. students, Melissa Gardner and Clarence Chan, are using computer modeling to predict and understand how cells respond to the mechanical stiffness of their environment and how chromosomes are segregated.
"A key challenge in stem cell research has been understanding how to direct stem cells into the various cell types that live in our bodies," said Odde. "The question is: How does a stem cell know what type of cell it should become?"
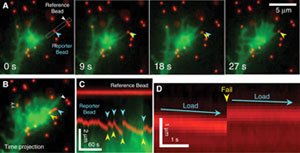
Using neurons as their test cells, the researchers found that cells sense their environment through the force it takes to pull against surrounding objects. Each cell uses its own internal skeleton and molecular motors, and the group found that the cell has an "optimum" stiffness where resistance to pulling is greatest. When cells are in an optimal stiffness environment, they found that the internal skeleton rhythmically plucks on its environment as if it were a guitar string. The results of their research are published in the Dec. 12 issue of "Science."
Odde's team also published findings about how replicated chromosomes are segregated. When cells divide, they must provide a complete set of chromosomes to each of the two "daughter" cells. Immediately prior to segregation the chromosomes align in a plate in the middle of the cell, a structure commonly seen in the microscope by researchers and students throughout the last century. Yet, the mechanism of chromosome centering has remained mysterious.
Using yeast cells, the team discovered that nanometer-scale molecular motors move along nanometer-scale cylinders called "microtubules," much like trains move on railroad tracks. Upon arrival at the end of the microtubules, the motors destabilize the microtubules to tear up the tracks along which they moved. They found that the longer the microtubule, the more motors there were on the microtubule ends, so that long microtubules tend to shorten and short microtubules tend to lengthen. This created two "clusters" of microtubule ends, one from each of two daughter cells. Since the chromosomes are attached to the clustered microtubule ends, the chromosomes end up clustered into two groups, and so become ready for separation into the daughter cells. The team developed a detailed computer model for how the chromosomes move, and their results were published in the Nov. 28 issue of the journal Cell.
Odde said it is important to understand the chromosome alignment process because misalignment during cell division plays an important role in how cancer progresses in the body.
"Both of these research findings have major implications for our understanding of tissue development, stem cells in regenerative medicine, and perhaps, even how cancer develops and metastasizes," Odde said. "By being able to make cells behave in a certain way, we can better control how stem cells replace injured body tissue when they are transplanted."
December 16, 2008
