David Odde and Paolo Provenzano: Predicting cancer cell movement
Biomedical engineering faculty study how tumor cells move in patients
March 25, 2018
Engineers in many fields develop mathematical models to simulate real-world events or predict future conditions. Two familiar examples are models to forecast tomorrow’s weather, and flight simulators to mimic the response of an airplane to the actions of a pilot at the controls.
Now two researchers in the College of Science and Engineering, who are also members of the Masonic Cancer Center, are using an $8.2 million Physical Sciences Oncology Center grant from the National Cancer Institute to develop a mathematical simulator. The simulator will be used to predict the movement of cancer through the human body, based on the physical characteristics of cancer cells and their surrounding micro-environment.
David Odde, professor of biomedical engineering, calls it a “cell migration simulator.”
The work will focus on tumors of the brain and pancreas, two especially lethal kinds of cancer. But the principals of what they learn should apply to other cancers as well.
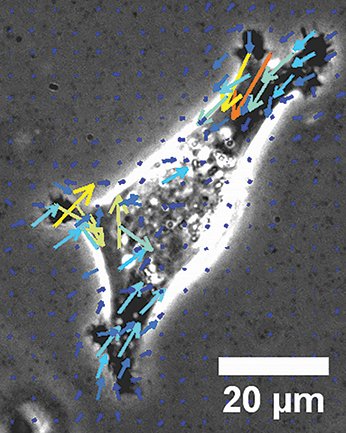
“We found that we get kind of surprising behaviors from a model for cell migration that allows us to predict cell behaviors in sometimes not-intuitive ways,” Odde said. “That I think will lead us to better strategies for treating cancer patients.”
Identifying patterns and tweaking the models
“In theory, you’d be able to turn different ‘knobs’—change the environment around the cell, change the levels of certain proteins within the cell, change the rate at which cells are sampling their environment,” said Paolo Provenzano, an assistant professor of biomedical engineering, who is Odde’s colleague on the project.
“As a consequence, you will be able to predict or tune how the cell would move and migrate,” said Provenzano.
The simulator will be based on the physics of the cancer cells and their immediate environment—real mechanical stuff like the force generation by the cell and the resistance offered by the environment.
The model, like those for everything from golf simulators to climate models, will be continually compared to results in real life and tweaked accordingly.
Tailor made for individuals
The researchers hope to tailor the model to individual patients, depending on the genetics of the patient and the cellular behavior of the tumor. Cancer patients at University clinics are being asked if they are willing to donate tissue samples for the research.
“So then we put some of these cancer cells into some of these environments to see how they behave and we can link that to patient data,” Provenzano said.
In their research, Odde is looking at behaviors and forces inside tumor cells. Provenzano is examining the exterior of the cells and the immediate environment—what he calls “the cell-environment interface.”
If the tumor were a car, “we kind of target the tires,” said Provenzano. “But then we also target the road.”
“Those physical properties really dictate how the disease behaves—whether a tumor grows, stalls, metastasizes, or switches back and forth between behaviors,” Provenzano explains. “We now know what dictates those decisions is largely this environment around the cell as a function of underlying genetics.”
Cancer cells are able to move using “muscles” called molecular myosin motors. Provenzano describes the movement as resembling an octopus pushing through small openings.
“Cells send out protrusions. They can push. They create these adhesions, they pull,” Provenzano said.
“There are times they ball up and squeeze through spaces,” he explained. “There are times they lay at like a person crawling on their elbows across the floor, and they switch through these phenotypes as necessary to navigate complex environments.”
The cells seem to move along well-established paths, which might be vulnerable to drugs.
Stopping cells on the highway
“Some of our recent work has shown one of the best ways to stop cancer cell movement might be to blow up the ‘highways’ where these cells travel,” Provenzano said.
Cancer cells also rely on their microenvironment as a barrier to drugs.
“Tumors are good at creating physical barriers to that process,” Provenzano said. “A huge part of what we’re trying to learn about is how to overcome that physical barrier.”
According to Odde, the greatest value of the simulator would be predicting which treatments, such as drug therapy, are most likely to be effective, and which are likely to be ineffective or even harmful.
“Without a model to guide that clinical strategy, we’re going to miss great opportunities,” he said.
Story by Greg Breining
Related stories
Efie Kokkoli: Tiny technology treats big disease
Chad Myers: Exploring genetic connections in breast cancer
If you’d like to support innovations within the College of Science and Engineering, visit the CSE giving web page.
