Biophysics Faculty
Faculty
J. Woods Halley
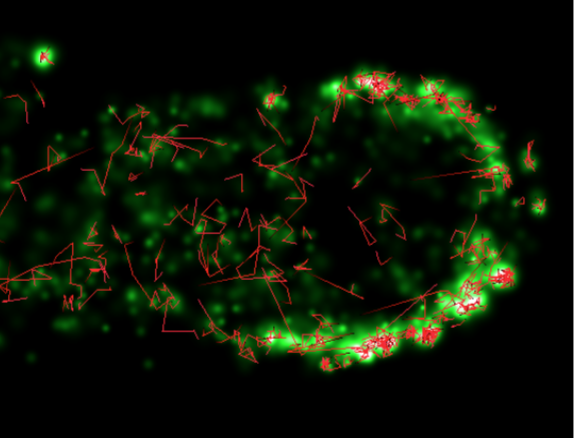
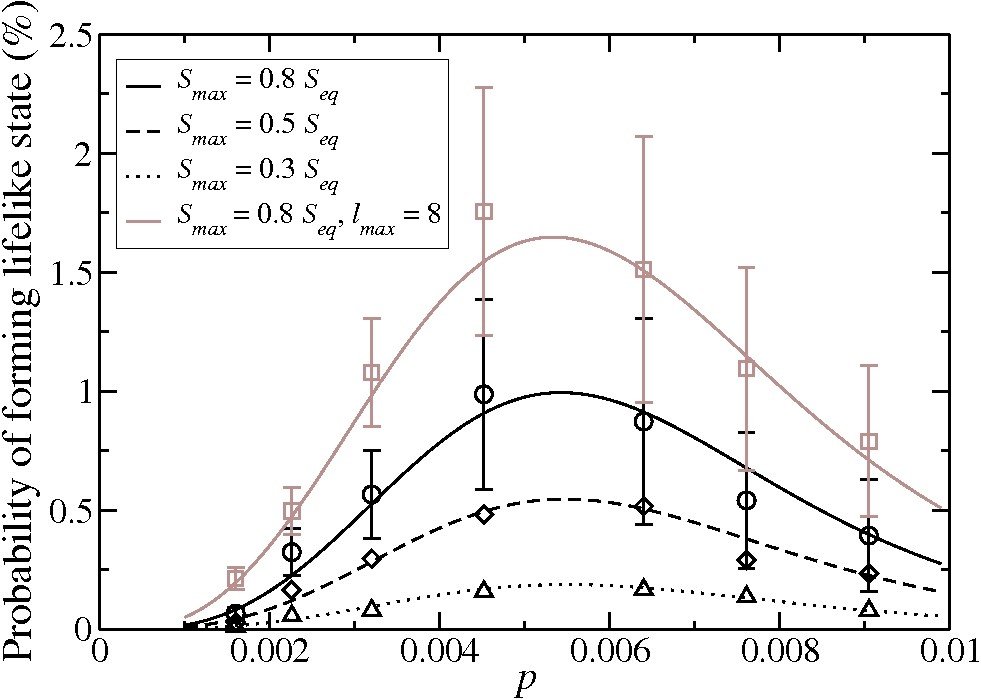
We study kinetic models for the formation of polymers far from equilibrium, which are intended to describe essential statistical features associated with the origin of life. The fundamental statistical problem is that, entropically, systems are overwhelmingly more likely to achieve chemical equilibrium than to be trapped in metastable states with lifelike characteristics. In the models we study, the kinetic trapping associated with such metastable states is induced by a parameter which reduces the fraction possible reactions which can occur. If that parameter (called p) is small enough, then kinetic trapping is observed to occur in numerical simulations, yielding a finite (though small) probability that metastable dynamic systems with some lifelike characteristics can arise. The figure shows the result for one such simulation. For very small p, no states at all of interest can arise, so we find a maximum probability at a finite value of p. Because that value of p is very small, we characterize this result informally as the idea that ’deserts may be better than ponds’ for inducing the initiation of life. We also study the effects of spatial diffusion, temperature and the number of monomer species on these conclusions, as well as the detailed nature of the metastable states that we find. Comparison of our model results with biological data on proteomes of prokaryotes shows that measures which we propose for detecting lifelike systems do in fact distinguish living from nonliving systems. Currently we focus on the effects of rapid quenches of solutions of prebiotic materials from high to low temperature on the likelihood of formation of lifelike systems. We find that quenching increases the likelihood and we hypothesize that, since such quenches have occurred continuously in ocean trenches for hundreds of millions of years, they may have played an important role in the origin of life. We are collaborating with experimentalist John Yin at the University of Wisconsin-Madison in some of this work.
Below: Numerical results for the probability of forming a lifelike state as a function of the parameter p describing the fraction of possible reactions which can occur. The fraction Smax/Seq measures how far the states allowed as lifelike are required to be from equilibrium. From Physical Review E 89, 022725 (2014).
Jochen Mueller
Cells are the elementary building blocks of all living matter. Despite their central role in carrying out all essential functions required to sustain life, our knowledge of how cells function on a molecular level remains rudimentary. Initially, a cell was viewed as a gigantic network of second-order differential equations, each equation representing collisional interactions between two biomolecules. However, it became apparent that the interactions between biomolecules within a cell are much more intricate than suggested by the above crude model. We now know that biomolecules assemble into larger complexes in order to carry out all essential cellular functions. These assemblies are not static, but dissociate or recruit additional members as a consequence of cellular signaling events and other environmental factors. This dynamic network of interactions is ultimately responsible for organizing the execution of all cellular processes. Our lab focuses on identifying these interactions by observing biomolecular assemblies directly inside the living cell. We developed and continue to develop new experimental approaches that quantify the spatiotemporal dynamics of interacting biomolecules. We also apply these methods to investigate specific cellular process, such as the assembly of retroviruses and mechanotransduction at the nuclear envelope.
Vincent Noireaux
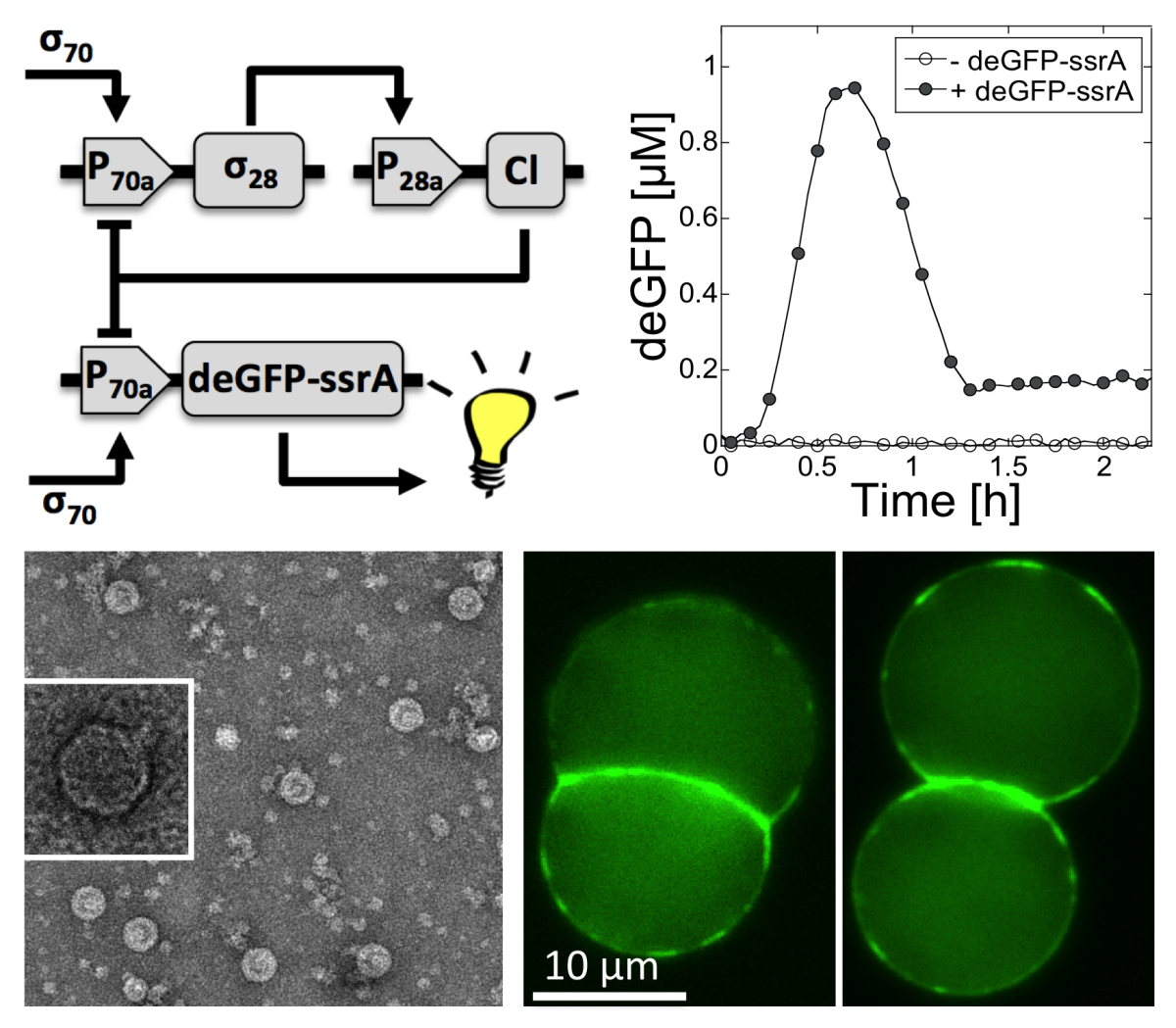
My lab has developed a cell-free transcription-translation (TX-TL) platform to construct and to study complex biochemical systems in vitro.
Unlike the other cell-free expression systems, our platform is an all E. coli extract that works with the endogenous TX and TL machineries. DNA programs (plasmids) are executed in a cell-free TX-TL mix entirely prepared in our lab. Our research includes: (I) the development and optimization of a cell-free TX-TL system for synthetic biology, (II) the construction and modeling of gene circuits in vitro, (III) the execution of large DNA programs such as bacteriophages, (IV) the bottom-up construction of an artificial cell. Our work is both fundamental and applied, with the goal of understanding the cooperative link between DNA information, self-assembly and metabolism, which covers the research areas of synthetic biology and biological physics.
Elias Puchner
Our lab employs and develops novel biophysical techniques to study cellular processes with high precision and detail.
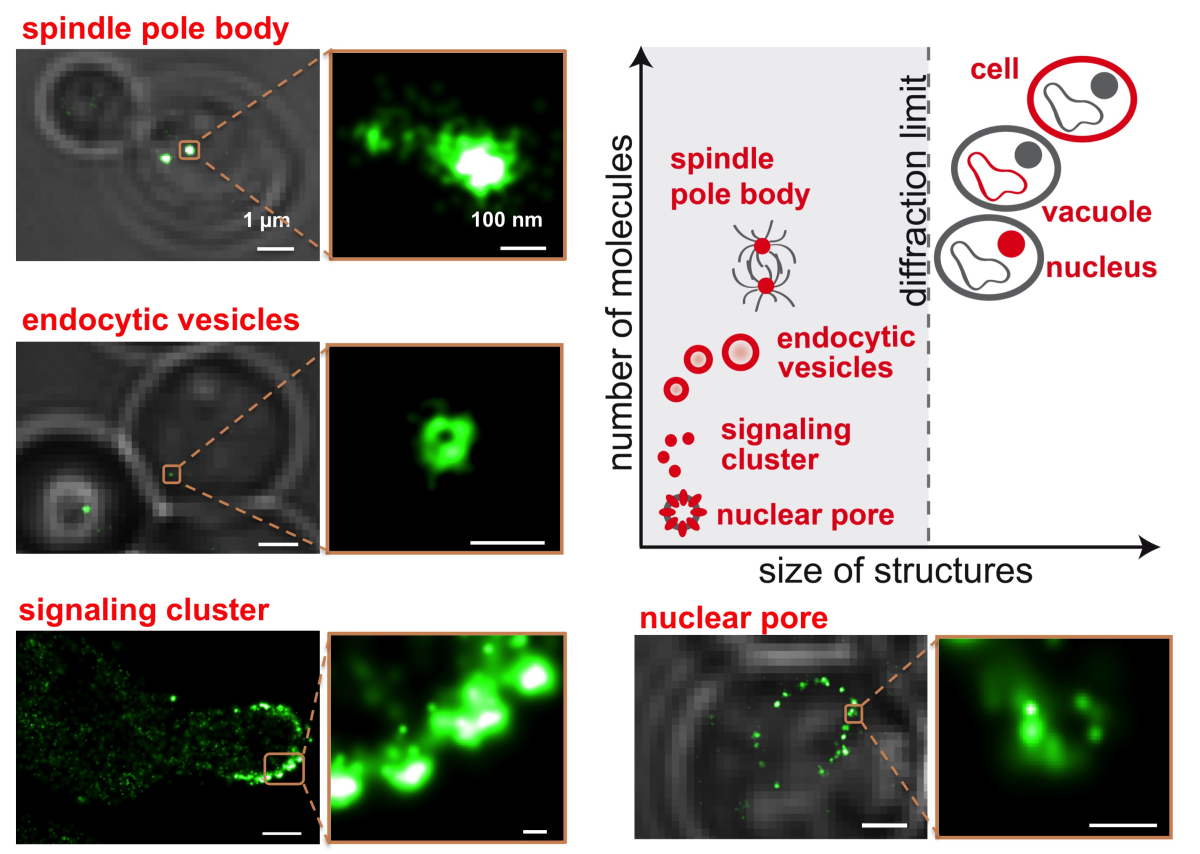
Living cells have the remarkable ability to sense external signals such as physical force or small molecules. This information is processed by intracellular signaling networks, which allow a cell to respond to the stimuli. Our research, at the intersection of physics and biology, aims to understand how these cellular signaling processes are regulated on different hierarchical length-scales by employing and further developing single molecule and super-resolution microscopy techniques.
Our goal is to investigate and characterize the biophysical principles of cellular signaling networks by connecting the behavior of single molecules and their complexes to the biological response of the entire cell. We approach this problem with a wide range of techniques – synthetic biology, genetic engineering, and molecular biology – but our specialty is quantitative single molecule super-resolution microscopy. This integrative approach allows us to precisely decipher the spatial organization and dynamics of biomolecules within cells below the optical diffraction limit, while simultaneously manipulating their localizations and interactions with tools from optogenetics and synthetic biology. By measuring the cell’s signaling activity, we can link the induced perturbations to regulation mechanisms in these signaling networks.
Associations:
Mayo Clinic, Graduate School of Biomedical Sciences
Jorge Viñals
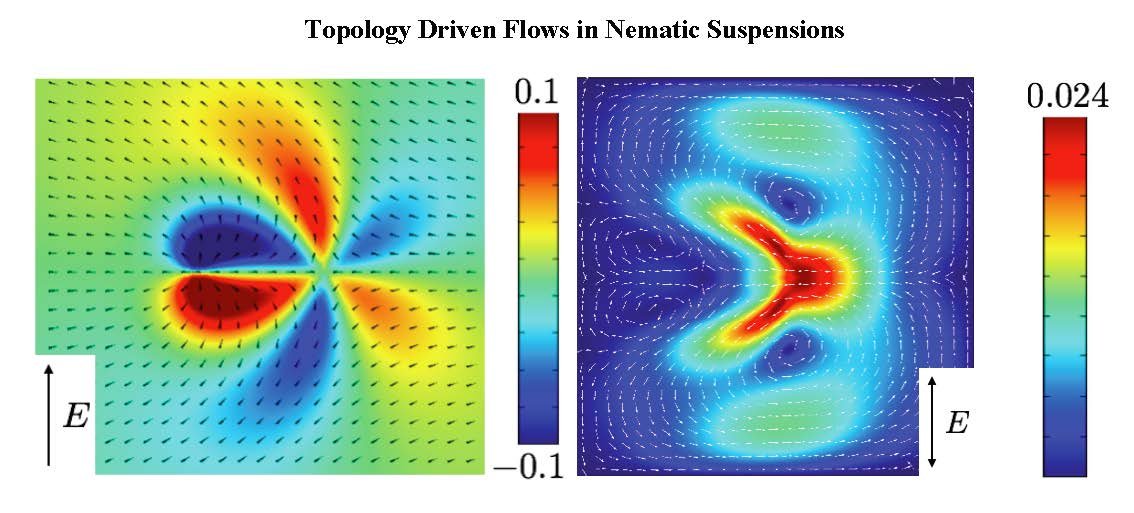
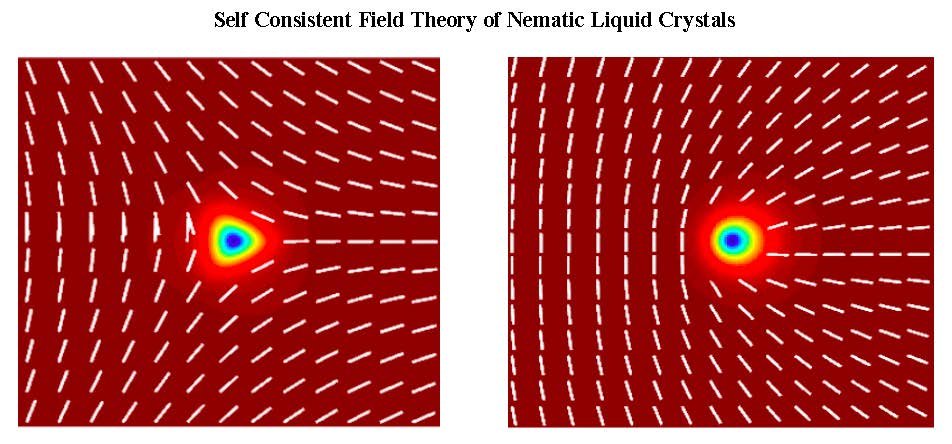
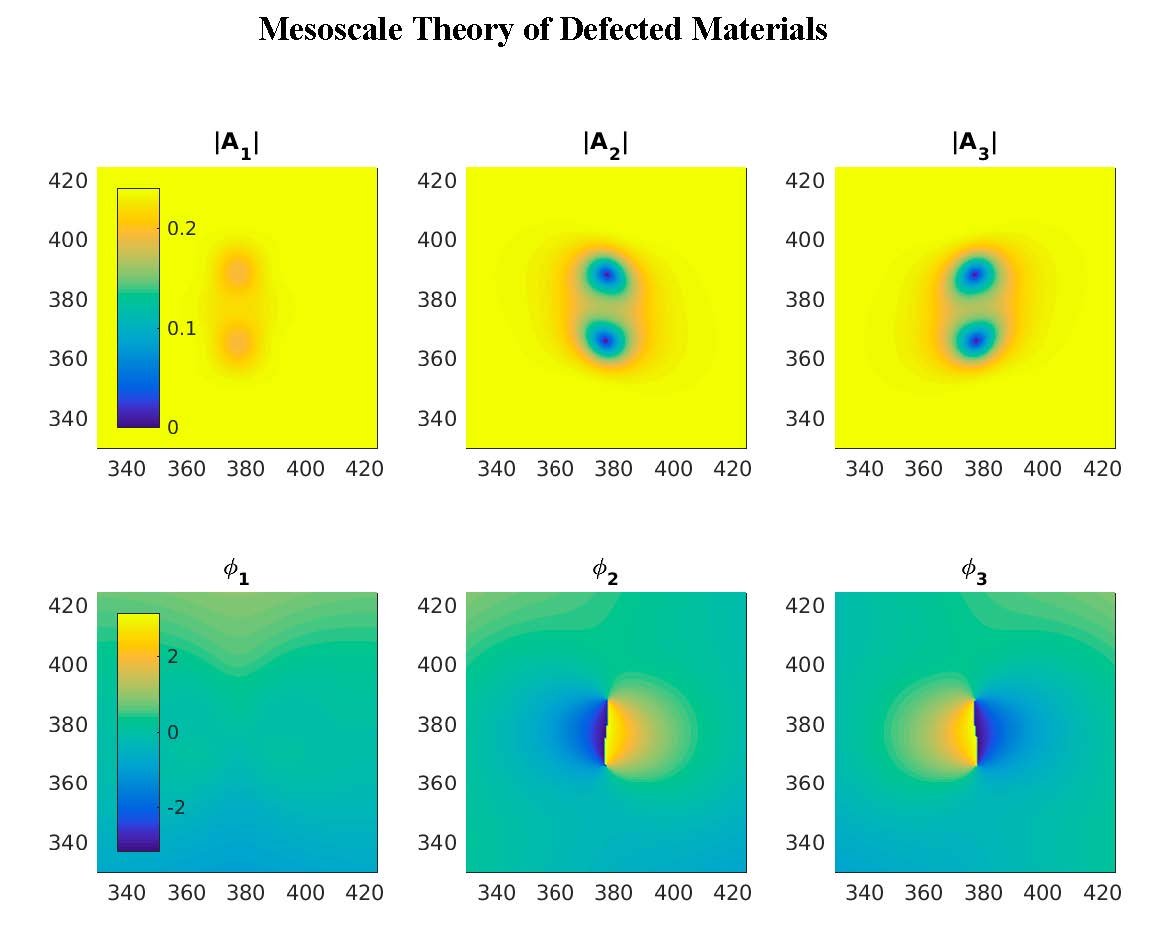
Our research is of theoretical and computational nature, and focuses on mesoscale theories of nonequilibrium phenomena in extended systems, with applications to Statistical Mechanics, Soft Matter Physics, and Materials Science. Generically, systems outside of thermodynamic equilibrium involve unstable interfaces and moving topological defects. We develop theories at the mesoscale of the relevant phases, analyze their macroscopic scale asymptotics, and obtain their non equilibrium properties through large scale computation. Our research is currently motivated by topologically driven flows in liquid crystals, unstable interfaces in complex fluids, and a mesoscale description of plasticity in defected materials.
Research is currently being done in these four areas:


Aaron Wynveen (Teaching Associate Professor)
My research can broadly be classified as soft matter theory and computation. Studies involve DNA and its interactions, e.g., how DNA can be condensed and packed within cells and viruses, how DNA forms various mesomorphic structures (in vivo and in vitro), and how DNA molecules recognize each other as a precursor to homologous recombination. More generically, I also study how molecules interact in solution, developing coarse grained models to describe large biomolecules. This research has been extended to study more specific systems, such as polymer and polyelectrolyte brushes. I have also been involved in studies of various molecular dynamics models, e.g., a self-consistent tight binding model of water, as well as modeling prebiotic systems to make predictions concerning the origins of life.
Emeriti
John Broadhurst (Emeritus)
I am working in two areas. The first is enhancing the effect of photon radiation in suppressing cancerous tissue. I am using the insertion of high atomic number compounds into tumors and irradiating with >1 Mev photons to generate energetic pairs of positrons and electrons. Also I am studying the brain response to sound. This uses magneto=encephalography to study the activity of brain cells after a sound of a known harmonic and time structure has arrived at the ear.