Professor William DeGrado
Paul G. Gassman Lectureship in Chemistry
Professor William DeGrado
Department of Pharmaceutical Chemistry
University of California San Francisco
Host: Professor Mark Distefano
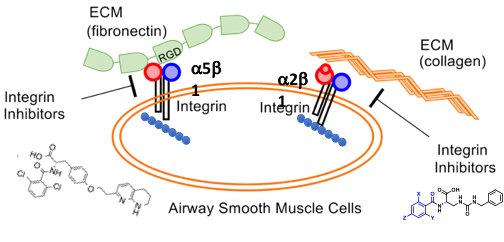
Use of integrin antagonists to disrupt pathological mechanical force-dependent processes in fibrosis and severe asthma.
Integrins are a class of dimeric membrane proteins, which connect extracellular protein ligands to cytoplasmic alpha-smooth muscle actin (aSMA). The force generated by aSMA contraction is transmitted through integrins to the bound extracellular ligands, playing an important in a number of biological and pathophysiological processes. For example, transforming growth factor beta is expressed as a latent precursor that is simultaneously bound to the extracellular matrix and one of a number of integrin subtypes, including avß1 and avß6. In stiff fibrotic tissue this results in activation of TGF-ß in an actin-dependent manner. This pathological feed-forward loop can be inhibited using small molecule inhibitors of the appropriate integrins. We are also examining the role of integrin-mediated force generation in the exaggerated airway narrowing and contraction observed in severe asthma. We are designing integrin antagonists to disrupt the actin-extracellular bridge between airway smooth muscle and the extracellular matrix. In animal models, these antagonists are able to decrease the transmission of force and they mitigate hyper-responsiveness, while leaving normal contractile responses intact.
Research
In the UC San Francisco lab of William DeGrado, PhD, we study the structural characterization of membrane proteins and de novo protein design in order to understand biological processes relevant to human disease and to develop novel therapeutics.
One primary research interest is de novo design, in which one designs proteins beginning from first principles. This approach critically tests our understanding of protein folding and function, while also laying the groundwork for the design of proteins and biomimetic polymers with properties not seen in nature.
De novo design of proteins has proven to be a useful approach for understanding the features in a protein sequence that cause it to fold into its unique three-dimensional structure. It has been possible to design functionally interesting proteins that bind redox-active cofactors, DNA, and transition metals. This approach has been extended to the design of membrane-active proteins, including ion channels, antibiotics, and fusogenic agents.
Professor William DeGrado
De novo protein design; membrane proteins; small molecule drug discovery for antimicrobials, influenza A virus, antifibrotics, and neurodegeneration; chemical biology; peptide design
