Computer simulator helps steer COVID-19 clinical trial toward promising treatment

August 18, 2022 — To help figure out which COVID-19 treatments might work best, a University of Minnesota clinical trial deployed a unique tool: a computer simulator.
The biophysics-based model simulated the disease on a molecular and cellular level so the trial team could screen potential treatments computationally long before they were given to participants.
The simulator informed the design of the clinical trial, the results of which were published in the New England Journal of Medicine yesterday. This trial, which was led by Carolyn Bramante, MD, MPH, an Assistant Professor at the University’s Medical School examined the efficacy of metformin, fluvoxamine and ivermectin to fight COVID-19.
“The trial is a good example of how engineering tools can be used to predict clinical outcomes,” says David Odde, a co-investigator of the trial and Professor in the Department of Biomedical Engineering who led the team that developed the simulator. “By steering research efforts toward more effective interventions and away from less effective ones, the COVID-19 simulator focused clinical trial efforts and ultimately helped add to the body of knowledge around disease treatments.”
Finding metformin effective
The trial was the nation’s first phase 3 study of whether metformin, a medication for type 2 diabetes; fluvoxamine, an antidepressant; and ivermectin, an antiparasitic, or their combinations could serve as possible treatments to prevent ER visits or hospitalization, as well as Long-COVID.
“Our trial suggests that metformin may reduce the likelihood of needing to go to the emergency room or be hospitalized for COVID-19,” said Dr. Bramante in a University news release. The randomized double-blinded placebo-controlled trial found that the diabetes drug lowered the odds of emergency department visits, hospitalizations, or death due to COVID-19 by over 40 percent; and over 50 percent if prescribed early in onset of symptoms.
Selecting a winner
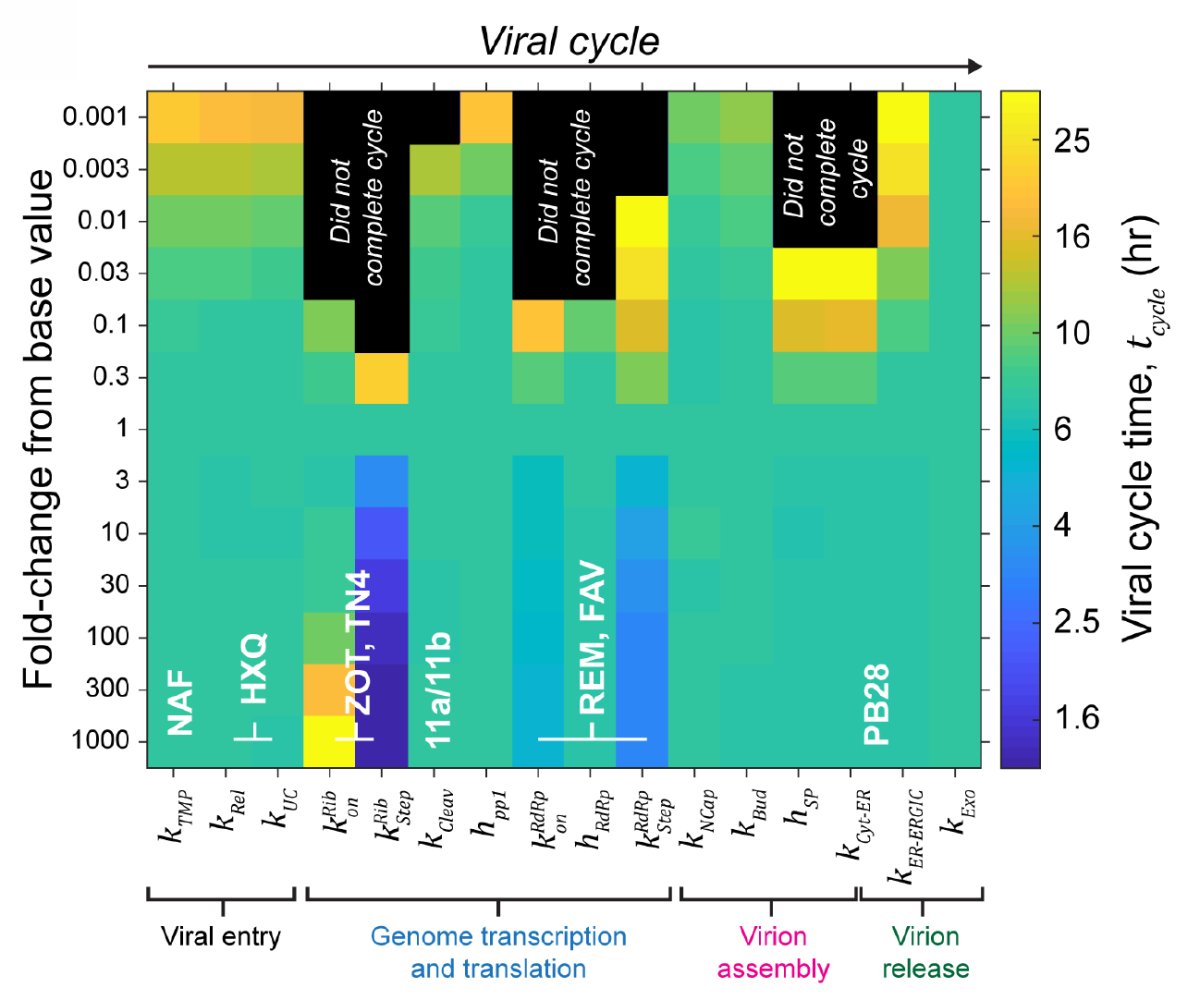
The COVID-19 simulator identified metformin’s strong potential in the early days of the pandemic. Developed with the help of UMN engineers, immunologists, and infectious disease experts, the tool simulates the life cycle of SARS-CoV-2 from infection of a healthy cell to release of progeny virions.
The simulator predicted that protein synthesis was a particularly good target for antiviral therapy, leading Prof. Odde and team to hypothesize that a drug capable of blocking this molecular process — like metformin — could be an effective tool for fighting the disease.
Meanwhile, other members of the clinical trial team noticed metformin’s potential through their own methods. In April 2020, natural language processing of SARS-CoV-1 and SARS-CoV-2 performed by Christopher Tignanelli, MD, and Rui Zhang, PhD, Director of the University’s Natural Language Processing/Information Extraction Program identified a class of drugs known as mTOR inhibitors — which includes metformin — that had strong potential to stop the viral life cycle.
Two independent methods both pointing toward metformin strengthened Dr. Bramante’s case for including the drug in the trial, and dovetailed with her own hypothesis that metformin’s immune effects could also help fight the disease.
Forecasting other outcomes
The simulator also predicted fluvoxamine would likely be effective (at a different dosage than studied in the trial), but did not make a prediction regarding ivermectin due to its uncertain mechanism of action.
The model has been highly accurate to date, successfully predicting, among others, the failure of hydroxychloroquine and the success of remdesivir before the results of clinical trials testing these therapies were announced.
One approach for many diseases
The clinical trial shows disease simulators could help bring new therapies into practice.
“More than 86 percent of all drug development programs fail,” said Prof. Odde, who also co-directs the Cancer Bioengineering Initiative. “In solid tumor oncology, where my research is focused, this failure rate exceeds 95 percent. These failures consume precious resources: dollars, clinical trial capacity, and the hopes of patients struggling to overcome disease. Computer simulators are a promising and relatively inexpensive way to reduce this failure rate and unearth more successful treatments.”
Creating disease simulators has long been a focus of Prof. Odde’s work, which has included building similar tools to predict the progression of cancer, sickle cell disease, and neurodegenerative disorders. Last fall, he received a $9.6 million P01 grant to pioneer a biophysics-based simulator for predicting tumor progression and ultimately accelerating new cancer immunotherapeutics.