Amanda Patsis is awarded UMII-MnDRIVE Graduate Assistantship
-Written by Declan Ramirez, Ph.D. Student
The MnDRIVE PhD Graduate Assistantship Program provides funding to PhD graduate students at the University of Minnesota (UMN) whose research involves the use of informatics to explore challenges faced by the state of Minnesota. This year, eleven PhD students at the UMN have been awarded PhD Graduate Assistantships through the UMN Informatics Institute (UMII), including Earth & Environmental Science’s own Amanda Patsis. Amanda’s work was funded under the MnDRIVE goal of Advancing Industry and Conserving Our Environment, due to its potential to aid in environmental conservation and bioremediation. Her project is entitled “Metagenome mining to inform bioremediation: Elucidating the impact of microbial metabolism on the transport fate of sulfur in Minnesota groundwater.”

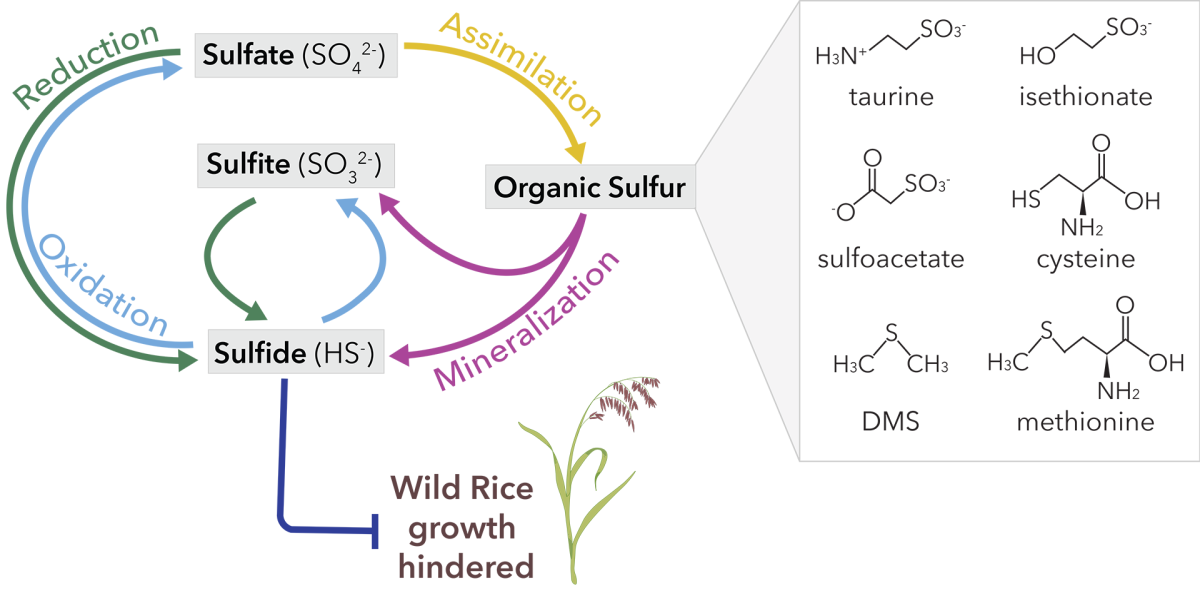
Amanda is a third year PhD student working in the Santelli Geomicrobiology Lab, where she investigates how microbes interact with and alter their environment. For her current PhD project, she is using publicly available datasets and her own data (collected from field work in the Soudan Underground Mine) to better constrain the impact of microbial communities on Minnesota’s sulfur cycle. Amanda explains that industrial activities like mining have led to the accumulation of excess sulfate in many of Minnesota’s surface waters, and that “microbial processes can produce, breakdown, or transform sulfur compounds, influencing how they move through and impact the environment.” For example, the production of sulfide (from sulfate) can hinder the growth of wild rice, an ecologically and culturally significant plant, while the incorporation of sulfur into organic compounds can remove it from surface waters.
Amanda’s MnDRIVE project will build on her current research, zooming out to look at a variety of different environments in Minnesota, including the sulfate-impacted Lakes Manganika and McQuade, a peatland bog, and the Soudan Underground Mine. Additionally, she’ll be using new and existing datasets to cut a broad swath across the state, looking for trends in microbial sulfur use that might be influencing surface water sulfate concentrations. Amanda explains that by understanding how microbes interact with different sulfur compounds “we can understand what that means for the fate of sulfur, its movement within a system, and if particular microbes are contributing to reducing excess sulfide in the environment.” This knowledge can guide bioremediation efforts by revealing which microbes to target in a given environment. Once identified, researchers can provide nutrients that help increase the abundance of microbes that remove harmful forms of sulfur, creating a natural solution to remediate surface water and groundwater.
Methodologically, much of Amanda’s work is on the computer where she compares microbial DNA sequences to databases of known genes. Identifying genes involved in sulfur cycling reveals if microbes have the ability to alter the form or transport fate of sulfur. However, at the Soudan Underground Mine Amanda collects her own samples, which involves filtering large quantities of water to capture microbes. Back at the lab Amanda ‘squishes’ the microbes, separates out their DNA, and runs it through a sequencer to get the sequence of bases in that DNA. Amanda says that one of the most interesting parts of the process is trying to reassemble the millions of DNA fragments that result. She likes to think of it as “doing 100 puzzles if someone mixed all the pieces together,” where each fragment of DNA is a puzzle piece that needs to be fitted together to recreate the original microbe. The really tricky part is that there’s more than one organism in the mix, so first she has to figure out which puzzle (microbe) the piece belongs to, and then how the pieces (DNA fragments) fit together. Luckily, computers help with this process! Once Amanda reconstructs the genome of a microbe she can look for different pathways that allow the microbes to use inorganic and organic sulfur, and try to identify particular genes that would allow them to create or remove harmful sulfur compounds from the environment.

Amanda’s favorite part of doing research is repurposing data that other groups have collected to inform her project. She states that “mining previously collected datasets for different types of genes is a very powerful tool that can help us answer new questions.” Overall, Amanda is very excited to receive this award. She has always been very interested in bioremediation but hasn’t been able to really focus on that aspect before in her work. With support from MnDRIVE Amanda says it's nice for that side to “be something that is driving the questions I ask and how I think about my results.” Amanda would like to thank her advisors Cara Santelli and Cody Sheik for their part in her earning this award.