Past Seminars & Events
Chao Sun
Thursday, Jan. 27, 2022, 9:45 a.m. through Thursday, Jan. 27, 2022, 11 a.m.
This seminar will be live-streamed
Zoom Link
Chao Sun, Ph.D.
Max Planck Institute for Brain Research
Frankfurt, Germany
Host: Professor Michael Bowser
Neuronal Protein Synthesis and Degradation Machines
A single neuron hosts ~10000 synapses in its complex dendritic and axonal arbour. Its protein logistics is run by amazing molecular machines put together with exquisite precision, such as ribosomes (protein-synthesis machines) and proteasomes (protein-degradation machines). Meanwhile, protein synthesis and degradation is essential for information storage in the brain. How do protein-synthesis & -degradation machines meet the huge demands of parallel processing by numerous synapses? To investigate this, I visualized protein -synthesis & -degradation machines as well as newly synthesized proteins near numerous synapses using quantitative, multiplexed, single-molecule localization microscopy. Combined with metabolic labelling, biochemistry, and synaptic-activity manipulations, these studies mapped and measured the subcellular protein-synthesis & -degradation capacity in complex neurons. The single-molecule resolution further affords intriguing insights into the subcellular adaptation of molecular machine assembly and function.
Chao Sun
Dr. Chao Sun is an EMBO & HFSP postdoctoral fellow in Prof. Erin M. Schuman's lab at the Max Planck Institute for Brain Research in Frankfurt, Germany. He uses quantitative, multiplexed super-resolution microscopy to investigate the molecular resource supply for the vast population of synapses associated with a single neuron. Chao obtained his PhD with Prof. William R. Dichtel at Cornell University (2013-2016) and later at Northwestern University (2016-2018). where he designed and manipulated molecular interactions to interface two-dimensional materials and molecular analogues for creating smart nano- materials and devices. His future research continues to focus on understanding and creating molecular systems that can 'think'.
Paul Ohno
Tuesday, Jan. 25, 2022, 9:45 a.m. through Tuesday, Jan. 25, 2022, 11 a.m.
This seminar will be presented 'in person' and live-streamed
331 Smith Hall
Zoom Link
Paul Ohno, Ph.D.
Schmidt Science Fellows
Environmental Fellow, Harvard University
Charged interface and atmospheric aerosol properties via spectroscopy
Chemical and physical processes at charged interfaces play a key role in environmental, biological, and technological systems. Yet, the relative scarcity of interface-selective experimental techniques has left open important questions regarding molecular-level structure and composition in a range of interfacial systems. Second harmonic generation (SHG) has become a workhorse interface-selective technique to characterize interfacial electrostatics. However, interpretation of SHG experiments has long been complicated by an inability to experimentally separate two terms that contribute to the total SHG signal. Here, I describe how the detection of not only SHG amplitude, but also its optical phase, enables separation and quantification of these two terms. The first term can then be used as an experimental benchmark for atomistic simulations of interfacial structure and composition, while the second term is proportional to interfacial potential and thus represents an “optical voltmeter”. I describe the design of an apparatus capable of measuring this optical phase from buried interfaces and demonstrate the application of this method to characterize oxide:water and lipid bilayer:water interfaces. Atmospheric aerosol particle phase state represents another property that has remained challenging to experimentally characterize due to the small size, low density, and delicate nature of the particles. Aerosol particles can undergo liquid-liquid phase separation (LLPS), impacting atmospheric processes including gas-particle partitioning, solar radiation scattering, and cloud nucleation. Here, I describe the first realization of an experimental technique capable of directly probing LLPS in particles of atmospherically-realistic sizes while they remain in suspension. Solvatochromic probe molecules are incorporated into the particles and their fluorescence emission is used to determine particle phase state. Differences in separation relative humidity (SRH) values measured here and previously reported SRH values from optical microscopy of much larger particles on substrates underscore the utility and importance of the technique and motivate future studies into the size dependence of LLPS in the atmosphere. Finally, I comment on the necessity of direct experimental characterizations of aerosol particle surfaces and interfaces to gain a comprehensive understanding of particle processes, properties, and impacts on air quality and climate.
Research
Paul’s overall scientific research interests revolve around the environment, sustainability, and chemistry. He is inspired by his childhood in the beautiful state of Maine and observations of the impacts of human activities on the natural environment around him.
Paul Ohno
Paul Ohno is a physical chemist studying the physical and chemical properties of secondary organic aerosol particles and the implications of these properties for the climate system.
Paul earned his AB in Chemistry from Princeton University in 2014 and his PhD in Chemistry from Northwestern University in 2019. During his PhD studies, he used laser spectroscopy to measure fundamental properties of aqueous interfaces so as to better understand, predict, and control chemical processes that occur there, like groundwater pollutant capture at the mineral/water interface.
As an Environmental Fellow, Paul works with Professor Scot Martin of the John A. Paulson School of Engineering and Applied Sciences and the Department of Earth and Planetary Sciences. Their work focuses on developing and applying spectroscopic techniques to directly determine physical and chemical properties, such as viscosity and diffusivity, of secondary organic aerosol particles while they remain in suspension. Paul is also a 2019 Schmidt Science Fellow.
Kevin D. Clark
Thursday, Jan. 20, 2022, 9:45 a.m. through Thursday, Jan. 20, 2022, 11 a.m.
This seminar will be presented 'in person' and live-streamed
331 Smith Hall
Zoom Link
Kevin D. Clark, Ph.D.
Beckman Institute
University of Illinois at Urbana-Champaign
Host: Professor Michael Bowser
Characterizing RNA Modifications in the Central Nervous System and Single Neurons:
Strategies in Sample Preparation and Mass Spectrometry
Apart from the four canonical nucleotides of RNA (A, U, G, and C), an expanding collection of over 150 RNA modifications exists that substantially diversifies the RNA code. Known as the “epitranscriptome,” these unusual chemical modifications to RNA range in complexity from simple methylations to conjugation with cellular metabolites and are enzymatically deposited/removed with sequence specificity to control the biophysical properties of cellular RNA biopolymers. However, characterizing RNA modifications in the central nervous system (CNS) has been limited by the complexity of animal models and lack of analytical methods capable of simultaneously detecting multiple modified RNAs in small-volume samples. These shortcomings have rendered the field of RNA modifications unable to account for the heterogeneous distribution of epitranscriptomic marks that potentially exists across the CNS and in single neurons. In this presentation, I will describe how our recent efforts to develop sample preparation and mass spectrometry approaches for the analysis of modified RNAs are beginning to reveal a previously uncharacterized link between dynamic RNA modifications and CNS function.
Kevin Clark
Kevin Clark received his Ph.D. in Analytical Chemistry from Iowa State University in 2018 and was awarded a 2018 Beckman Institute Postdoctoral Fellowship to study RNA modifications in the central nervous system (CNS) with Prof. Jonathan Sweedler and Prof. Rhanor Gillette. Now a postdoctoral research associate in Prof. Sweedler’s lab, Dr. Clark is developing separations and mass spectrometry methods for profiling RNA modifications in single neurons to better understand how they contribute to CNS function.
Tian (Autumn) Qiu
Tuesday, Jan. 18, 2022, 9:45 a.m. through Tuesday, Jan. 18, 2022, 11 a.m.
This seminar will be presented 'in person' and live-streamed
331 Smith Hall
Zoom Link
Tian (Autumn) Qiu, Ph.D.
Beckman Institute
University of Illinois at Urbana-Champaign
Host: Professor Michael Bowser
Exploring Molecular Mechanisms of Nano-Bio and Host-Microbe Interactions
The dynamics between the environment, host and microbes have huge implications for both sustainability and human health. Understanding the molecular basis of how microbiota impact the host response to environmental exposure are critical to develop effective strategies for risk assessment, pollution control and precision medicines. In this seminar, I will introduce my previous work on nano-bio and host-microbe interactions, building towards my future research program to decipher the molecular basis of the environment-host-microbe interactions. The rapidly advancing field of nanotechnology poses challenges for timely and predictive risk assessment of highly reactive nanomaterials being released into the environment. Focusing on an emerging nanoscale battery material, we investigated DNA damage as a shared toxicity mechanism in two bacteria and revealed unprecedented details on DNA modification after nanomaterial exposure as well as biological responses relating to DNA damage. Expanding my research to the host-microbe interactions, my current work investigates the role of D-amino acids, particularly D-alanine (D-Ala), as microbe-originated signaling molecules in the microbiome-gut-brain axis. Using nematode Caenorhabditis elegans and Escherichia coli bacterial mutants, I found impact of bacterial D-Ala synthesis and metabolism on animal phenotypes under normal condition and high glucose exposure. Using germ-free mice, I revealed biodistribution and sources of D-Ala in higher animals. As my previous studies have provided new biomarkers for nanomaterial risk assessment and indicated new targets for microbiota-mediated intervention of diseases, my future research will combine my previous trainings to investigate the molecular basis of the environment-host-microbe interactions.
Tian (Autumn) Qiu
Dr. Tian (Autumn) Qiu completed her BS in Chemistry at Peking University in 2012 and PhD in Chemistry at University of Minnesota in 2018. Her PhD work focused on understanding nanotoxicity mechanisms to environmental bacteria at the molecular level with bioanalytical tools in the laboratory of Prof. Christy Haynes as part of the Center for Sustainable Nanotechnology. From 2018 to 2021, she was a Beckman Postodctoral Fellow at University of Illinois, Urbana-Champaign, working with Prof. Jonathan Sweedler and other collaborators on exploring role of D-amino acids in the microbiome-gut-brain axis using animal and microbial models. She is currently continuing her work as a Postdoctoral Research Associate in the Sweedler Lab.
Kaiyu Fu
Thursday, Jan. 13, 2022, 9:45 a.m. through Thursday, Jan. 13, 2022, 11 a.m.
This seminar will be presented 'in person' and live-streamed
331 Smith Hall
Zoom Link
Kaiyu Fu
Department of Radiology and Electrical Engineering
Stanford University
Host: Professor Michael Bowser
In vivo Real-time Biosensor: Convergence of Electroanalytical Chemistry, Nanotechnology and Bioengineering
The emerging biosensing devices largely extend our capability to measure and monitor the biomolecules in living organisms. Yet most commercially available detection platforms are ether simplified for a handful of abundant molecules in vivo, e.g., glucose and electrolytes, or designed with time-consuming pretreatment procedures for targets with a lower concentration. There is an urgent quest to integrate the advancement of analytical chemistry and nanotechnology to solve the long-existing problems for biodetection.
In this talk, I will first present a novel electrochemical detection platform that consists of high-density nanoscale electrodes, viz. nanopore electrode array (NEA), for ultrasensitive biomolecule detection. Through a robust fabrication method to achieve nanopores with embedded electrodes, I will explore how electron transfer and ion transport are coupled in the nanostructures for the m\t (ass-limited samples. Next, I will present a real-time sensing technology to measure biomolecules in live animals based on the above-mentioned nanoporous electrode. In the first example, I will discuss an implantable NEA drug sensor to collect tissue-based pharmacokinetics. So far, the efficacy and safety of chemotherapy are measured based on blood samples. This NEA drug sensor can continuously measure the concentrations of the chemotherapy drug at multiple tumor sites in a rodent model. Due to the tumor microenvironment heterogeneity, it demonstrates apparent differences in pharmacokinetics relative to the circulation that could meaningfully affect treatment. Last, I will introduce the second example that an implantable NEA neurochemical sensor can continuously measure the neurotransmitters in the cerebrospinal fluid of the brain. This NEA neurochemical sensor is designed on a flexible substrate with multiple sensing elements for chronic recording at different brain regions. In addition, the whole system is integrated with a wireless module as a potentially cable-free device to track the behaviors of freely moving animals associated with the detection of neurochemical signals. Overall, this talk will depict a generalized approach to access in vivo real-time biochemical information for future clinical applications.
Research
My research overlaps three disciplines: electrochemistry, nanoscience, and synthetic biology. Several emerging and exciting topics derived from these fields surge upon instrumentation advancement, cutting-edge nanotechnology, and commercial biomedical devices over the past decade. Recently, I am particularly interested in 1) nanoscale electrochemistry, which paves the way to the development of ultrasensitive analytical platforms reaching a single-entity detection level; 2) electrochemical biosensors for point-of-care diagnostics and chronic disease management; and 3) nanobiotechnology, e.g., DNA nanotechnology and direction evolution of nucleic acids, aiming to manipulate the molecular recognition with unprecedented precision and efficiency.
Kaiyu Fu
Kaiyu Fu is a postdoctoral associate of Radiology and Electrical Engineering at Stanford. He obtained his Ph.D. degree in Analytical Chemistry from Notre Dame. Kaiyu is one of the ACS Division of Analytical Chemistry Graduate Fellowship recipients. He is broadly interested in advancing electroanalytical methods, including the fabrication of micro/nanoscale electrodes for lab-chip devices and the application of electrochemical biosensors for point-of-care diagnostics.
Varun Gadkari
Tuesday, Jan. 11, 2022, 9:45 a.m. through Tuesday, Jan. 11, 2022, 11 a.m.
This seminar will be presented 'in person' and live-streamed
331 Smith Hall
Zoom Link
Varun Gadkari
Department of Chemistry
University of Michigan
Host: Professor Michael Bowser
Applications of Mass Spectrometry Beyond Mass Measurement: Characterizing Biomolecular Structure in the Gas Phase
Rapid advances in structural biology have enabled the visualization of three- dimensional biomolecular structures yielding valuable insights into biological structure-function relationships. However, due to the stringent sample quantity/quality requirements of current structure elucidation techniques, some essential biomolecular classes have been excluded from characterization. New structural biology techniques are necessary which are capable of analyzing these intractable samples and elucidating their structural properties. In this seminar, I will present my work aimed at developing mass spectrometry-based methods for the characterization of challenging biological targets which elude conventional structural biology. This includes development of specialized instrumentation and methods for gas phase biomolecular measurements, and the application of these approaches to characterize various biologically challenging systems such as neurodegenerative disease-linked amyloidogenic proteins, near-megadalton protein complexes, and nucleic acids. In total, my work highlights the benefits of combining bioanalytical chemistry, chemical biology, and structural biology to characterize otherwise inaccessible biomolecules, and provides a path forward for the investigation of other biological questions in the absence of conventional structural data.
Research
I am a postdoctoral research fellow at the University of Michigan developing and applying native mass spectrometry techniques to bridge the gap between biochemistry and analytical chemistry. My long-term goal is to develop mass spectrometry based approaches for the study of degenerative diseases. I earned my Doctor of Philosophy (Ph.D.) in Biochemistry at The Ohio State University where I characterized various DNA replication pathways by biophysical and biochemical techniques.
Varun Gadkari
Varun Gadkari obtained his Bachelor of Science degree in biochemistry from The Ohio State University in 2012. He then joined the Ohio State Biochemistry Program to pursue his graduate studies in the laboratory of Prof. Zucai Suo, where he used biochemical and biophysical approaches to characterize DNA Replication/Repair proteins. Upon completing his doctorate in biochemistry in 2017, Varun became a postdoctoral research fellow in the laboratory of Prof. Brandon Ruotolo at the University of Michigan where he currently develops native mass spectrometry methods for gas phase structural biology, with a focus on characterizing protein complexes, amyloidogenic proteins, and nucleic acids.
Professor William DeGrado
Thursday, Dec. 2, 2021, 9:45 a.m. through Thursday, Dec. 2, 2021, 11 a.m.
This seminar will be presented remotely and live-streamed
331 Smith Hall
Zoom Link
Paul G. Gassman Lectureship in Chemistry
Professor William DeGrado
Department of Pharmaceutical Chemistry
University of California San Francisco
Host: Professor Mark Distefano
Analysis and design of proton transporters
The mechanism of proton transport through membrane proteins is of general interest to multiple areas of biology. In enveloped RNA viruses, viroporins (viral proton channels) facilitate the transfer of protons through the viral envelope to disrupt protein-RNA interactions required for uncoating, as well as control of the pH of cellular compartments. Using a variety of spectroscopic, crystallographic, and computational methods, we have investigated the mechanism by which protons are conducted through the M2 proton channel from influenza A virus, and used this information to design new anti-influenza drugs that target highly drug-resistant forms of the virus.
A second topic of the talk will focus on the use of de novo protein design to test the principles underlying membrane protein folding, stability and dynamics. We have also used this knowledge to design membrane proteins that test the fundamental mechanisms underlying selective transport of protons, and linking proton movement to transport of other ions.
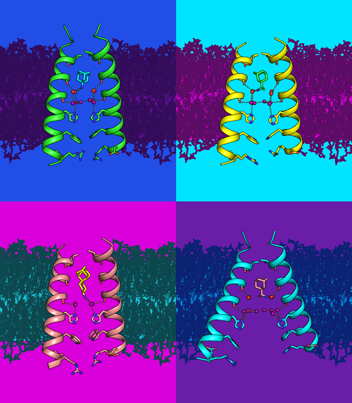
Research
In the UC San Francisco lab of William DeGrado, PhD, we study the structural characterization of membrane proteins and de novo protein design in order to understand biological processes relevant to human disease and to develop novel therapeutics.
One primary research interest is de novo design, in which one designs proteins beginning from first principles. This approach critically tests our understanding of protein folding and function, while also laying the groundwork for the design of proteins and biomimetic polymers with properties not seen in nature.
De novo design of proteins has proven to be a useful approach for understanding the features in a protein sequence that cause it to fold into its unique three-dimensional structure. It has been possible to design functionally interesting proteins that bind redox-active cofactors, DNA, and transition metals. This approach has been extended to the design of membrane-active proteins, including ion channels, antibiotics, and fusogenic agents.
Professor William DeGrado
De novo protein design; membrane proteins; small molecule drug discovery for antimicrobials, influenza A virus, antifibrotics, and neurodegeneration; chemical biology; peptide design
Professor William DeGrado
Tuesday, Nov. 30, 2021, 9:45 a.m. through Tuesday, Nov. 30, 2021, 11 a.m.
This seminar will be presented remotely and live-streamed
331 Smith Hall
Zoom Link
Paul G. Gassman Lectureship in Chemistry
Professor William DeGrado
Department of Pharmaceutical Chemistry
University of California San Francisco
Host: Professor Mark Distefano
De novo protein design
De novo protein design, in which one designs proteins beginning from first principles, provides an approach that critically tests our understanding of protein folding and function, while also laying the groundwork for the design of novel proteins and biomimetic polymers. The de novo design of proteins has proven to be a useful approach for understanding the features in a protein sequence that allow them to fold into their unique three-dimensional structures, as well as how their structures achieve complex functions from molecular recognition to catalysis. This talk will focus on the design of water-soluble proteins with predetermined structures and functions, including binding of metal ion cofactors and small molecules.
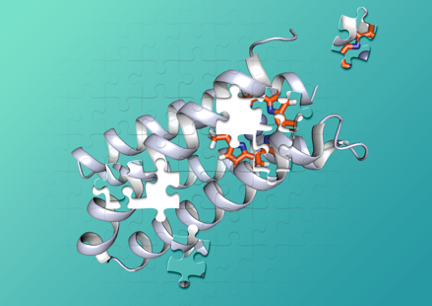
Research
In the UC San Francisco lab of William DeGrado, PhD, we study the structural characterization of membrane proteins and de novo protein design in order to understand biological processes relevant to human disease and to develop novel therapeutics.
One primary research interest is de novo design, in which one designs proteins beginning from first principles. This approach critically tests our understanding of protein folding and function, while also laying the groundwork for the design of proteins and biomimetic polymers with properties not seen in nature.
De novo design of proteins has proven to be a useful approach for understanding the features in a protein sequence that cause it to fold into its unique three-dimensional structure. It has been possible to design functionally interesting proteins that bind redox-active cofactors, DNA, and transition metals. This approach has been extended to the design of membrane-active proteins, including ion channels, antibiotics, and fusogenic agents.
Professor William DeGrado
De novo protein design; membrane proteins; small molecule drug discovery for antimicrobials, influenza A virus, antifibrotics, and neurodegeneration; chemical biology; peptide design
Professor William DeGrado
Monday, Nov. 29, 2021, 4 p.m. through Monday, Nov. 29, 2021, 5 p.m.
This seminar will be presented remotely and live-streamed
331 Smith Hall
Zoom Link
Paul G. Gassman Lectureship in Chemistry
Professor William DeGrado
Department of Pharmaceutical Chemistry
University of California San Francisco
Host: Professor Mark Distefano
Use of integrin antagonists to disrupt pathological mechanical force-dependent processes in fibrosis and severe asthma.
Integrins are a class of dimeric membrane proteins, which connect extracellular protein ligands to cytoplasmic alpha-smooth muscle actin (aSMA). The force generated by aSMA contraction is transmitted through integrins to the bound extracellular ligands, playing an important in a number of biological and pathophysiological processes. For example, transforming growth factor beta is expressed as a latent precursor that is simultaneously bound to the extracellular matrix and one of a number of integrin subtypes, including avß1 and avß6. In stiff fibrotic tissue this results in activation of TGF-ß in an actin-dependent manner. This pathological feed-forward loop can be inhibited using small molecule inhibitors of the appropriate integrins. We are also examining the role of integrin-mediated force generation in the exaggerated airway narrowing and contraction observed in severe asthma. We are designing integrin antagonists to disrupt the actin-extracellular bridge between airway smooth muscle and the extracellular matrix. In animal models, these antagonists are able to decrease the transmission of force and they mitigate hyper-responsiveness, while leaving normal contractile responses intact.
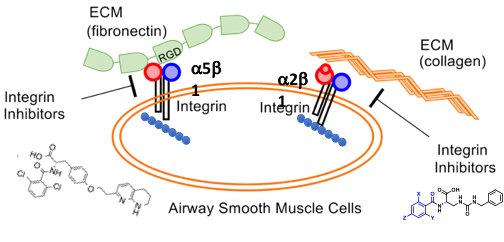
Research
In the UC San Francisco lab of William DeGrado, PhD, we study the structural characterization of membrane proteins and de novo protein design in order to understand biological processes relevant to human disease and to develop novel therapeutics.
One primary research interest is de novo design, in which one designs proteins beginning from first principles. This approach critically tests our understanding of protein folding and function, while also laying the groundwork for the design of proteins and biomimetic polymers with properties not seen in nature.
De novo design of proteins has proven to be a useful approach for understanding the features in a protein sequence that cause it to fold into its unique three-dimensional structure. It has been possible to design functionally interesting proteins that bind redox-active cofactors, DNA, and transition metals. This approach has been extended to the design of membrane-active proteins, including ion channels, antibiotics, and fusogenic agents.
Professor William DeGrado
De novo protein design; membrane proteins; small molecule drug discovery for antimicrobials, influenza A virus, antifibrotics, and neurodegeneration; chemical biology; peptide design
2nd Annual Sigma Aldrich Chemistry Symposium
Friday, Nov. 19, 2021, 9:45 a.m. through Friday, Nov. 19, 2021, 2 p.m.
Online
To attend this event, please pre-register using the link below:
http://bit.ly/midwest-chemistry21
2nd annual Midwest Sigma Aldrich Chemistry Symposium
Please join us Friday, November 19th, 2021 for the 2nd annual Midwest Sigma Aldrich Chemistry Symposium, where we will be hearing presentations from the 2021 Aldrich Distinguished Lecturers, Dr. Alison Wendlandt, Ph.D., Assistant Professor, Massachusetts Institute of Technology (Chosen by the University of Wisconsin-Madison) and Dr. Korey Carter, Ph.D, Assistant Professor, University of Iowa (chosen by the University of Iowa). Full Schedule.
The event will also include several short presentations from Midwest graduate student and post-doctoral researchers, as well as a talk from Dr. Jane Murray, Ph.D., Head of Global Green Chemistry, Merck KGaA, Darmstadt, Germany.
Agenda (all times CST):
9:45 AM - Opening - MilliporeSigma
10:00 AM - Selective Catalytic Isomerization Reactions - Dr. Alison Wendlandt, Ph.D., Massachusetts Institute of Technology (Hosted by the University of Wisconsin-Madison, Department of Chemistry)
11:00 AM - Cyrene and Cyrene Blends in Research - Dr. Jane Murray, Ph.D. -MilliporeSigma
11:30 AM - Combining Structural, Spectroscopic, and Biochemical Tools to Explore Coordination Chemistry Across the Actinide Series - Dr. Korey Carter, Ph.D., University of Iowa (Hosted by the University of Iowa, Department of Chemistry)
12:30 PM - Midwest Researcher Presentations
To attend this event, please pre-register using the link below:
http://bit.ly/midwest-chemistry21