Fighting COVID-19 with deep graphs
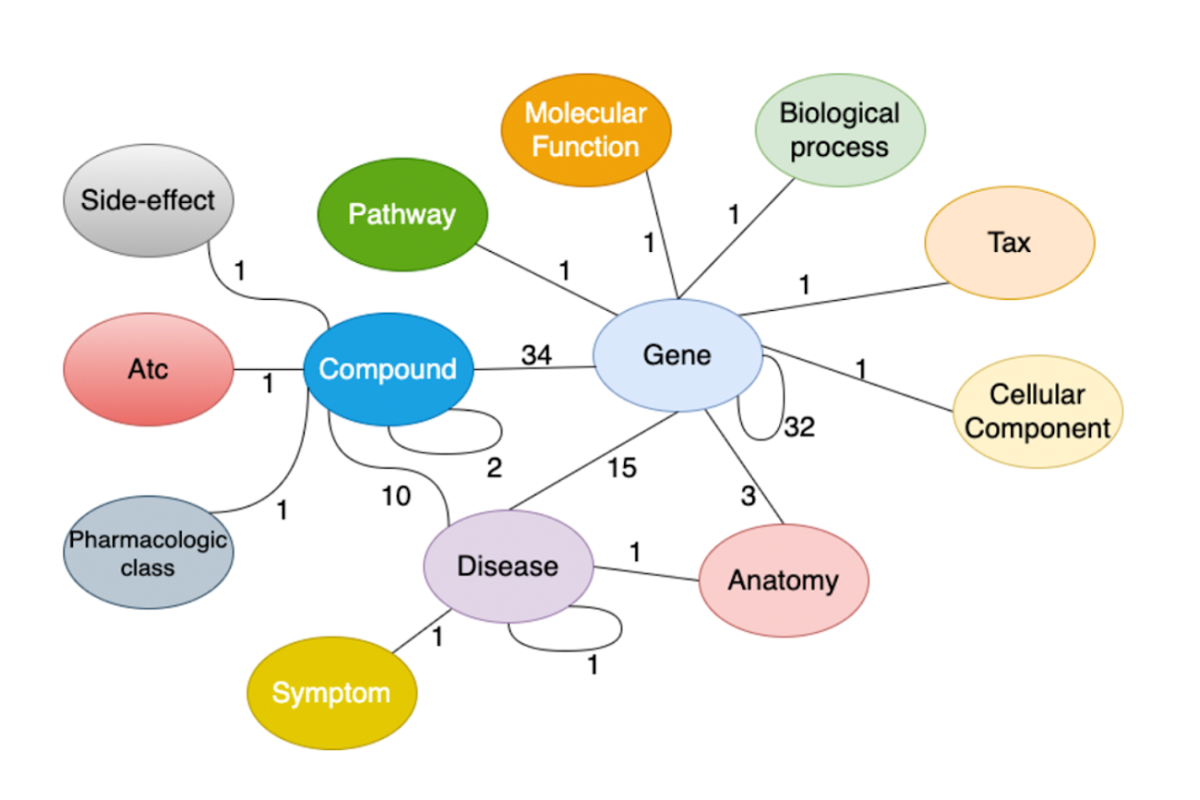
The rapid progress of the COVID-19 pandemic demonstrates the dire need for quick and effective drug discovery. Drug-repurposing is a drug discovery paradigm that uses existing drugs for new therapeutic indications. It has the advantages of significantly reducing the time and cost compared to de novo drug discovery. Drug-repurposing represents a promising strategy for COVID-19 treatment.
CS&E Professor George Karypis with Ph.D. students Vasileios Ioannidis (ECE) and Saurav Manchanda (CS&E) from the University of Minnesota, a team of scientists from the Ohio State University, Hunan University, and Amazon’s AWS AI labs in Shanghai and Palo Alto have created the Drug Repurposing Knowledge Graph (DRKG) and a set of machine learning tools that can be used to prioritize drugs for repurposing studies.
The graph shows how each component is linked and reveals potential interactions. Pre-existing drugs are included to determine whether they may be able to treat other diseases.
The goal of the graph is to provide other researchers with a list of drugs that will narrow their search, reduce time and save money.
“The basic idea of drug repurposing is you’re trying to find interactions among, essentially, drugs and diseases. Or, if you want to be a little bit more biological about it, you want to find interactions among drugs and genes that are associated with diseases.” said Ioannidis.
Each disease is associated with a set of genes, he said, with viruses encoded in the genes. Drugs typically relate to the gene in some way, like inhibiting a gene by preventing that gene from expressing another one.
“By following this high-level idea, we can follow this knowledge graph that if a drug is used to inhibit a certain gene and this gene has been found to be related to COVID-19 … then we can have some kind of prediction,” Ioannidis said.
Professor Karypis has been working on using computational methods like knowledge graphs for solving drug discovery problems for the last 15 years.
By using a knowledge graph to predict missing links and find incomplete information, researchers can discover potential treatments for diseases, he said.
“People can just use the knowledge graph and the sample code provided so they can do their own studies,” Karypis said.
The researchers are still working on the knowledge graph, which can be used for diseases beyond COVID-19. In the future, Karypis said they hope to develop a new set of methods that will help people to better leverage the drug knowledge graph for drug repurposing.
“At the end of the day, can we reduce [the] time to essentially ending the pandemic?” he said.
The DRKG includes, curates, and normalizes information from six publicly available databases and data that were collected from recent publications related to COVID-19. It has 97,238 entities belonging to 13 types of entities, and 5,874,261 triplets belonging to 107 types of relations.
The machine learning tools use the state-of-the-art deep graph learning methods (DGL-KE) to compute embeddings of DRKG entities and relations, and use these embeddings to predict how likely a drug can treat a disease or how likely a drug can bind to a protein associated with the disease. When tested against the human proteins associated with COVID-19, these tools identified with high scores many of the COVID-19 drug candidates that are currently under clinical trials.
The team has made DRKG publicly available on github along with a set of machine learning tools and pre-computed embeddings. This will enable researchers to conduct computational drug repurposing more efficiently and effectively for COVID-19 and for other diseases (e.g., Alzheimer’s disease).